If it comes to speak about the future possibilities of molecular biology, it is worth keeping an eye on the medical applications. And if you think about the most peculiar infection agents you should for sure think of viruses also. What is a virus? Of course we all know what a virus might cause to us, like a simple respiratory infection. These are usually caused by viral infections that later are super infected with bacteria. I had a professor who tried to explain us what a virus is. It skips almost any definitions. We can not be sure if we could consider a living thing at all!!! At the end he told us in a laconic way: a virus is a VIRUS! Nothing more.
If we try to find them it is good to know, that they were discovered through the observation that you can transfer an infection from one cell culture to the other even after filtrating the solution through a filter with 0.4 micrometer holes. That means that no bacteria can bypass this filter, but infections can be transferred with this solution. The firs experiments were done in order to monitor these infections, to see that after infection there was a "clean window period", a period when the infections agent disappeared from the cell culture. After this window the virus reappeared and the supernatant solution had infections properties again.
Today we know plenty of details about viruses. There are basically two flavours of them DNA and RNA viruses. So that is an important point! because we have plenty of molecular biology tools that allow us to characterize nucleic acids. One of the most complex tool from this series is the DNA microarray. As one of my students pointed out last week, in the next video from TED, we can have a wonderful presentation about how these tools can be used in a fast and relatively easy way to get a deeper insight in the world of viruses. As a perspective the video shows us some excellent diagnostic applications that will be probably used to develop state of the art diagnostic tools in the next couple of years.
So, let us see how it works!
More info about the viruses and vaccination, here.
Mar 30, 2009
Mar 15, 2009
Restriction Enzymes
Restriction enzymes are used to cut plasmids. We have tackled the plasmids in the previous lecture. You can have a full description about the restriction enzymes here.
As a most basic introduction I would say that restriction enzymes are enzymes of the bacteria representing a kind of immune function of the bacteria. They are present in pairs in bacteria: a DNA methylase and a restriction enzyme. They both recognize the same sequence. The bacteria is methylating its own DNA in a sequence specific manner. By this its own DNA is protected against any foreign DNA. Since horizontal gene transfer is quite common in bacteria, the bacterial cell can protect its own genetic material with the help of the restriction enzymes. The foreign DNA entering into the cell will present a different DNA methylation pattern. The unmethylated recognition sites will be cut by the restriction enzymes and by this destroyed.
Different bacterial species have different restriction enzymes with different recognition sites (certainly each has a DNA methyltransferase, too). The nomenclature of the restriction enzyme reflects their origin. In the most trivial case the name Eco RI enzyme is informing us that it has been isolated from Escherichia coli strain R and it has been the first to have been isolated from this strain.
In the molecular biology lab we use them to cut and manipulate plasmids. They are like scissors that can be directed to specific sites in the plasmid to cleave it. With an appropriate collection of site specific cutting enzymes we can step into the very exciting field of genetic engineering.
Let us have a look to some basic usage of restriction enzymes:
You can check a good introductory video here.
In any case when working with enzymes, please use latex gloves, and keep enzymes on ice!
The unit of a restriction enzyme "U" stands for the amount of enzyme needed to cut 1microgram of plasmid with a single cutting site, in one hour, in ideal environment.
The environment of the reaction is provided by buffers. The enzymes are usually provided in a concentration of 10U/ul (10 units per microliter). The enzymes are supplied in glicerol solution and always stored at -20 C. The buffer may as well come in a 10 fold concentrated solution (10X) and it should also be kept frozen.
A typical restriction enzyme reaction is set up in the following way:
1. Check the map of the plasmid for the distribution of the cutting sites.
2. Measure the concentration of the plasmid solution by spectrophotometer. Your plasmid concentration should be in the range of 1 microgram per microliter.
3. Calculate the volume of the plasmid needed to have the required amount of product at the end. The volume of the reaction should be kept as low as possible, and should not exceed 100 ul/ reaction tube. Use sterile, DNAse free microcentrifuge (so called) "Eppendorf" tubes.
4. Plan the reaction. You should have approx 1 to 10 U of enzyme per microgram of plasmid. In the final volume of the reaction the total volume of the enzyme should be less the 1/10, because higher glicerol concentration might alter the specificity of the reaction. The buffer will be 1/10 of the final volume. Keep the final volume low (less then 100 microliters). If needed, adjust the reaction volume to the planned final volume with nuclease free water. Check the optimal temperature for the reaction. It is usually 37C, but it might differ. Check for possible star activity of the enzyme in its data sheet.
Example:
Mix the following components (ul stands for microliter):
16ul Nuclease Free Water+
1ul Plasmid solution (concentration 1ug/ul)+
2ul 10X Buffer+
1ul Restriction Enzyme (10U/ul)
Total: 20ul
5. Once the reaction is planned, start to do it: bring ice, prepare tubes, melt the buffer in your hands.
6. Pipette the required volumes of water, plasmid and buffer into the tube.
7. Add the enzyme to the tube and mix gently. Do not vortex!
8. Put the reaction into the thermostat set to the required temperature.
9. Put the enzyme and the buffer back to -20C and clean up you bench!
10. After the allocated time has passed, stop the reaction. We are usually keeping the reaction in the thermostat for 4 hours. You can stop the reaction in several ways: by adding EDTA; by heat inactivating the enzyme at 85C for 10 minutes, or simply by freezing the tube and keeping it frozen until you purify it.
You can have a look on the applications in the video below.
Good luck!
As a most basic introduction I would say that restriction enzymes are enzymes of the bacteria representing a kind of immune function of the bacteria. They are present in pairs in bacteria: a DNA methylase and a restriction enzyme. They both recognize the same sequence. The bacteria is methylating its own DNA in a sequence specific manner. By this its own DNA is protected against any foreign DNA. Since horizontal gene transfer is quite common in bacteria, the bacterial cell can protect its own genetic material with the help of the restriction enzymes. The foreign DNA entering into the cell will present a different DNA methylation pattern. The unmethylated recognition sites will be cut by the restriction enzymes and by this destroyed.
Different bacterial species have different restriction enzymes with different recognition sites (certainly each has a DNA methyltransferase, too). The nomenclature of the restriction enzyme reflects their origin. In the most trivial case the name Eco RI enzyme is informing us that it has been isolated from Escherichia coli strain R and it has been the first to have been isolated from this strain.
In the molecular biology lab we use them to cut and manipulate plasmids. They are like scissors that can be directed to specific sites in the plasmid to cleave it. With an appropriate collection of site specific cutting enzymes we can step into the very exciting field of genetic engineering.
Let us have a look to some basic usage of restriction enzymes:
You can check a good introductory video here.
In any case when working with enzymes, please use latex gloves, and keep enzymes on ice!
The unit of a restriction enzyme "U" stands for the amount of enzyme needed to cut 1microgram of plasmid with a single cutting site, in one hour, in ideal environment.
The environment of the reaction is provided by buffers. The enzymes are usually provided in a concentration of 10U/ul (10 units per microliter). The enzymes are supplied in glicerol solution and always stored at -20 C. The buffer may as well come in a 10 fold concentrated solution (10X) and it should also be kept frozen.
A typical restriction enzyme reaction is set up in the following way:
1. Check the map of the plasmid for the distribution of the cutting sites.
2. Measure the concentration of the plasmid solution by spectrophotometer. Your plasmid concentration should be in the range of 1 microgram per microliter.
3. Calculate the volume of the plasmid needed to have the required amount of product at the end. The volume of the reaction should be kept as low as possible, and should not exceed 100 ul/ reaction tube. Use sterile, DNAse free microcentrifuge (so called) "Eppendorf" tubes.
4. Plan the reaction. You should have approx 1 to 10 U of enzyme per microgram of plasmid. In the final volume of the reaction the total volume of the enzyme should be less the 1/10, because higher glicerol concentration might alter the specificity of the reaction. The buffer will be 1/10 of the final volume. Keep the final volume low (less then 100 microliters). If needed, adjust the reaction volume to the planned final volume with nuclease free water. Check the optimal temperature for the reaction. It is usually 37C, but it might differ. Check for possible star activity of the enzyme in its data sheet.
Example:
Mix the following components (ul stands for microliter):
16ul Nuclease Free Water+
1ul Plasmid solution (concentration 1ug/ul)+
2ul 10X Buffer+
1ul Restriction Enzyme (10U/ul)
Total: 20ul
5. Once the reaction is planned, start to do it: bring ice, prepare tubes, melt the buffer in your hands.
6. Pipette the required volumes of water, plasmid and buffer into the tube.
7. Add the enzyme to the tube and mix gently. Do not vortex!
8. Put the reaction into the thermostat set to the required temperature.
9. Put the enzyme and the buffer back to -20C and clean up you bench!
10. After the allocated time has passed, stop the reaction. We are usually keeping the reaction in the thermostat for 4 hours. You can stop the reaction in several ways: by adding EDTA; by heat inactivating the enzyme at 85C for 10 minutes, or simply by freezing the tube and keeping it frozen until you purify it.
You can have a look on the applications in the video below.
Good luck!
Labels:
DNA,
Lab,
molecular biology,
plasmid,
Restriction Enzyme,
transfection,
Water
Mar 7, 2009
Green Fluorescent Protein or GFP
Green lights in the dark
When someone first shows up in our lab, the prime goal I set up for him or her is to make "green cells" - I mean to introduce a Green Fluorescent Protein into a mammalian cell culture. In order to be able to perform this one has to know some basic molecular biology. One has to know what a cell is, what the difference is between a prokaryote and an eukaryote cell; what the central dogma is namelly that the information flows from DNA to RNA and from here to proteins is, or as it has been formulated originally and still correctly, the information flows from nucleic acids towards proteins (albeit I assume we will see exceptions for this rule, too). (You can reach a very good lecture on this topic here.) One has to know what the difference between DNA and RNA is, in most basic approach the chemical difference is minuscule (there is a deoxyribose in the backbone of the DNA and a ribose in the RNA, there are other differences but this is the most prominent), while the results are spectacular. DNA is a quite stable molecule that can be degraded by DNAses. DNases require divalent metal ions for their activity ( usually Mg, but other divalent ions can be used too), and we can remove these ions from solutions with so called chelating agents. Most commonly we use EDTA for this task.
From practical point of view, one needs to have some backgrounds in order not to be lost in a molecular biology lab as it follows:
One has to be able to use pipettes (as seen in the previous posts), to make buffers, to know about pH, know what molarity is, and have a good basic background in maths (just enough to calculate the compositions of the buffers).
But you can perform the most basic experiment of DNA isolation even in the kitchen! At the end of this experiment you will be able to even SEE the DNA!
You can extract DNA from any cell, but the easiest way is to use some germs, like wheat or bean germs, soya germs and so on... In the following video you can see the procedure. If you do not have isopropyl alcohol (I don't have at home for example) use regular ethanol or a strong spirit with at least 70% alcohol content!
Regarding RNA, the world of RNA is a transient world. RNA is degraded by enzymes that can be found everywhere. RNAses can not be blocked by removing metal ions with EDTA. This makes the half life of RNA very short. Let us take the analology of the computer: DNA is like the information on the hard disk, one might have a software on the computer without using it- this is the information in the DNA. If one double clicks on its icon, the program starts, this corresponds to the transcription: information is transcribed from DNA to RNA, or the software is running, even if it is not yet in use, it is ready to get an input and process it into the output. The RNA is similarly translated by ribosome into proteins: these are the products that have been coded in the DNA. Or according to the computer analogy you create a document with the word processor software. The document is an entity by itself. You can print it and have it. If you turn off your computer, the temporary files are destroyed, all unsaved files are deleted. So is with the RNA. RNA is carrying an information for a short period of time, it has a short half life, but can be regenerated from the DNA. These processes are explained in the following video:
Ok, so how do we make green cells? Green flourescent protein is encoded in the genome of the Jelly fish. The protein once identified can be introduced into other organisms if we isolate the DNA sequence that is encoding the GFP protein. So let's have a look to these wonderful organisms!
Beautiful Jelly fish
The discovery of GFP protein and their mode of action changed plenty of studies in biology. The Nobel Prize for Chemistry in 2008 was given for the identification of the GFP protein and its way of action. You can see below two videos about the topic. A detailed, in depth one or below a short overview of the topic. You choose!
Giving green light to biology
Nobel Prize for GFP
After this overview I think it is time to have an experiment. We will see how you can introduce the GFP encoding DNA into a bacteria. For this we use so called plasmids as a vector. We call vector in biology a tool that is able to carry genetic information, like a plasmid, cosmid, or a virus. A plasmid is a small circular DNA that is able to self-replicate into a bacteria and to express a protein. They are responsible for lateral gene transfer in bacteria, e.g. transfering antibiotic resistance gene from one bacteria to a different one.
In the following experiment we will see the introduction of a GFP encoding DNA into a so called Agrobacterium, a bacteria that is infecting plants.
Introducing the GFP into a bacteria
Cool, isn't it?
We can make even more complicated investigations with the help of the GFP. In the following animation it is shown the transfection process in a mammalian cell where the addressed question is if two proteins interact or not? For this they use the so called FRET or fluorescence resonance energy transfer. In order to see if the two proteins are close to each other or not, we have to use two GFP like tagged proteins with their excitation and emission wave lengths close to each other. See how it works:
Investigating protein-protein interactions with fluorescent proteins
GFP has several other applications, like tracing of migrating neurons, as seen in the following video:
Or full GFP organisms like in the following one:
If you would like to know even more about the GFP protein, please visit the best site in this topic I have ever seen, the page of Marc Zimmer, here.
I think we had even too much of GFP now, so in the next posts we will go back to plasmids...
See you!
When someone first shows up in our lab, the prime goal I set up for him or her is to make "green cells" - I mean to introduce a Green Fluorescent Protein into a mammalian cell culture. In order to be able to perform this one has to know some basic molecular biology. One has to know what a cell is, what the difference is between a prokaryote and an eukaryote cell; what the central dogma is namelly that the information flows from DNA to RNA and from here to proteins is, or as it has been formulated originally and still correctly, the information flows from nucleic acids towards proteins (albeit I assume we will see exceptions for this rule, too). (You can reach a very good lecture on this topic here.) One has to know what the difference between DNA and RNA is, in most basic approach the chemical difference is minuscule (there is a deoxyribose in the backbone of the DNA and a ribose in the RNA, there are other differences but this is the most prominent), while the results are spectacular. DNA is a quite stable molecule that can be degraded by DNAses. DNases require divalent metal ions for their activity ( usually Mg, but other divalent ions can be used too), and we can remove these ions from solutions with so called chelating agents. Most commonly we use EDTA for this task.
From practical point of view, one needs to have some backgrounds in order not to be lost in a molecular biology lab as it follows:
One has to be able to use pipettes (as seen in the previous posts), to make buffers, to know about pH, know what molarity is, and have a good basic background in maths (just enough to calculate the compositions of the buffers).
But you can perform the most basic experiment of DNA isolation even in the kitchen! At the end of this experiment you will be able to even SEE the DNA!
You can extract DNA from any cell, but the easiest way is to use some germs, like wheat or bean germs, soya germs and so on... In the following video you can see the procedure. If you do not have isopropyl alcohol (I don't have at home for example) use regular ethanol or a strong spirit with at least 70% alcohol content!
Regarding RNA, the world of RNA is a transient world. RNA is degraded by enzymes that can be found everywhere. RNAses can not be blocked by removing metal ions with EDTA. This makes the half life of RNA very short. Let us take the analology of the computer: DNA is like the information on the hard disk, one might have a software on the computer without using it- this is the information in the DNA. If one double clicks on its icon, the program starts, this corresponds to the transcription: information is transcribed from DNA to RNA, or the software is running, even if it is not yet in use, it is ready to get an input and process it into the output. The RNA is similarly translated by ribosome into proteins: these are the products that have been coded in the DNA. Or according to the computer analogy you create a document with the word processor software. The document is an entity by itself. You can print it and have it. If you turn off your computer, the temporary files are destroyed, all unsaved files are deleted. So is with the RNA. RNA is carrying an information for a short period of time, it has a short half life, but can be regenerated from the DNA. These processes are explained in the following video:
Ok, so how do we make green cells? Green flourescent protein is encoded in the genome of the Jelly fish. The protein once identified can be introduced into other organisms if we isolate the DNA sequence that is encoding the GFP protein. So let's have a look to these wonderful organisms!
Beautiful Jelly fish
The discovery of GFP protein and their mode of action changed plenty of studies in biology. The Nobel Prize for Chemistry in 2008 was given for the identification of the GFP protein and its way of action. You can see below two videos about the topic. A detailed, in depth one or below a short overview of the topic. You choose!
Giving green light to biology
Nobel Prize for GFP
After this overview I think it is time to have an experiment. We will see how you can introduce the GFP encoding DNA into a bacteria. For this we use so called plasmids as a vector. We call vector in biology a tool that is able to carry genetic information, like a plasmid, cosmid, or a virus. A plasmid is a small circular DNA that is able to self-replicate into a bacteria and to express a protein. They are responsible for lateral gene transfer in bacteria, e.g. transfering antibiotic resistance gene from one bacteria to a different one.
In the following experiment we will see the introduction of a GFP encoding DNA into a so called Agrobacterium, a bacteria that is infecting plants.
Introducing the GFP into a bacteria
Cool, isn't it?
We can make even more complicated investigations with the help of the GFP. In the following animation it is shown the transfection process in a mammalian cell where the addressed question is if two proteins interact or not? For this they use the so called FRET or fluorescence resonance energy transfer. In order to see if the two proteins are close to each other or not, we have to use two GFP like tagged proteins with their excitation and emission wave lengths close to each other. See how it works:
Investigating protein-protein interactions with fluorescent proteins
GFP has several other applications, like tracing of migrating neurons, as seen in the following video:
Or full GFP organisms like in the following one:
If you would like to know even more about the GFP protein, please visit the best site in this topic I have ever seen, the page of Marc Zimmer, here.
I think we had even too much of GFP now, so in the next posts we will go back to plasmids...
See you!
Labels:
animation,
cell,
Central Dogma,
DNA,
DNA isolation,
DNAses,
EDTA,
FRET,
GFP,
Green fluorescent protein,
Jellyfish,
molecular biology,
plasmid,
RNA,
Transcription,
transfection,
Translation
Mar 2, 2009
Using Serological Pipettes
Dear Colleagues,
I promised you to give an update about serological pipettes.
We use serological pipettes when we want to manipulate (to move) liquids that are in the range of 5 to 25 ml. Smaller volumes than 5ml can be measured with Gilson type pipettes, while for larger volumes than 25ml we use measuring cylinders.
Serological pipettes have a dispensable graduated tube, and a filter that is not allowing contamination with any particles from the air. The pipettes look like this for example:
[caption id="attachment_199" align="aligncenter" width="468" caption="serological-pipette"]
 [/caption]
[/caption]They can be charged, have a button to move liquids up and one for release the aspirated liquids.
We use sterile, single packed pipettes in three different ranges: up to 5ml, to 10ml and to 25ml. They have different color codes:
[caption id="attachment_201" align="aligncenter" width="468" caption="sterile-serological-pipettes"]
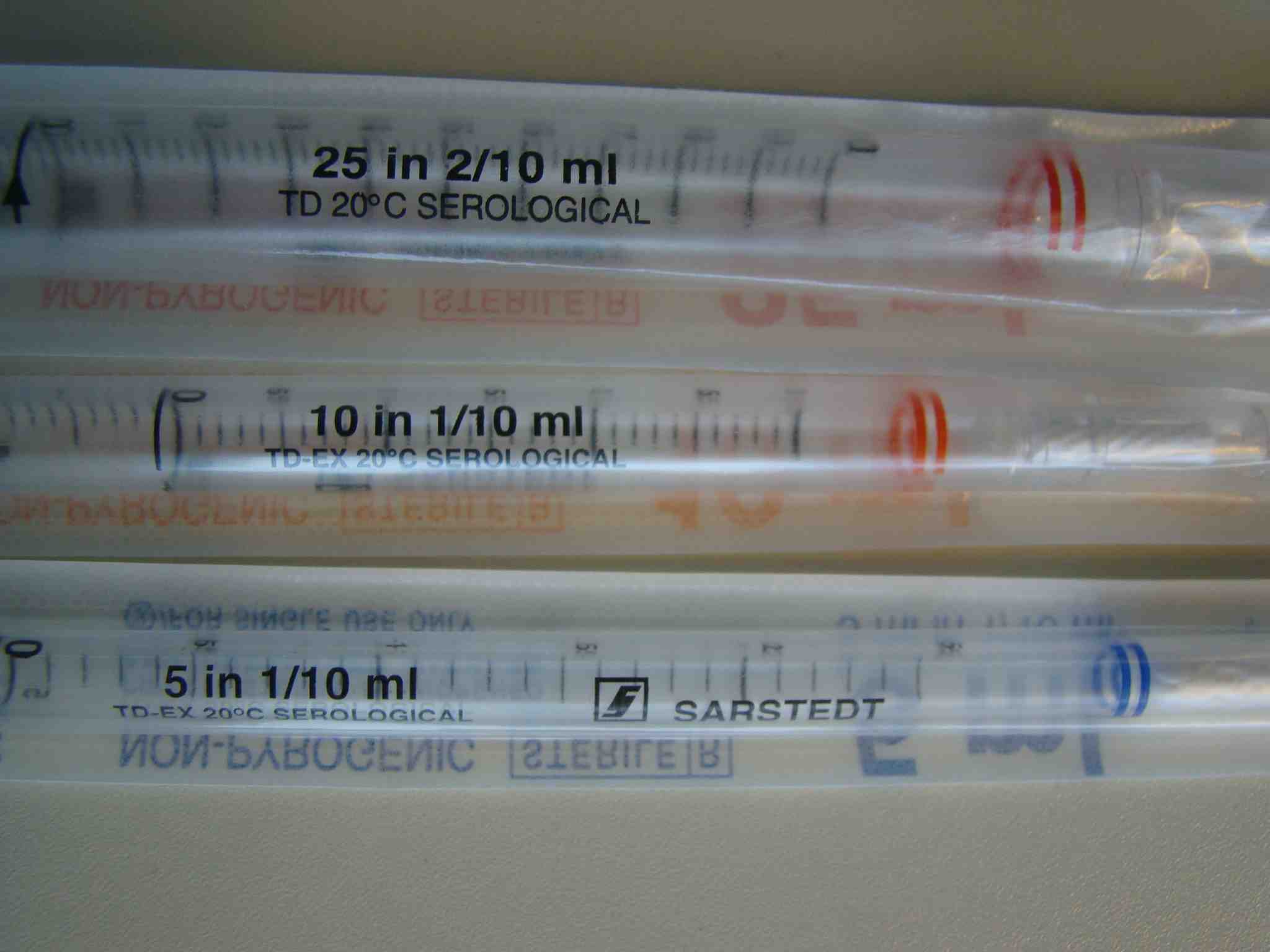 [/caption]
[/caption]The same from their back:
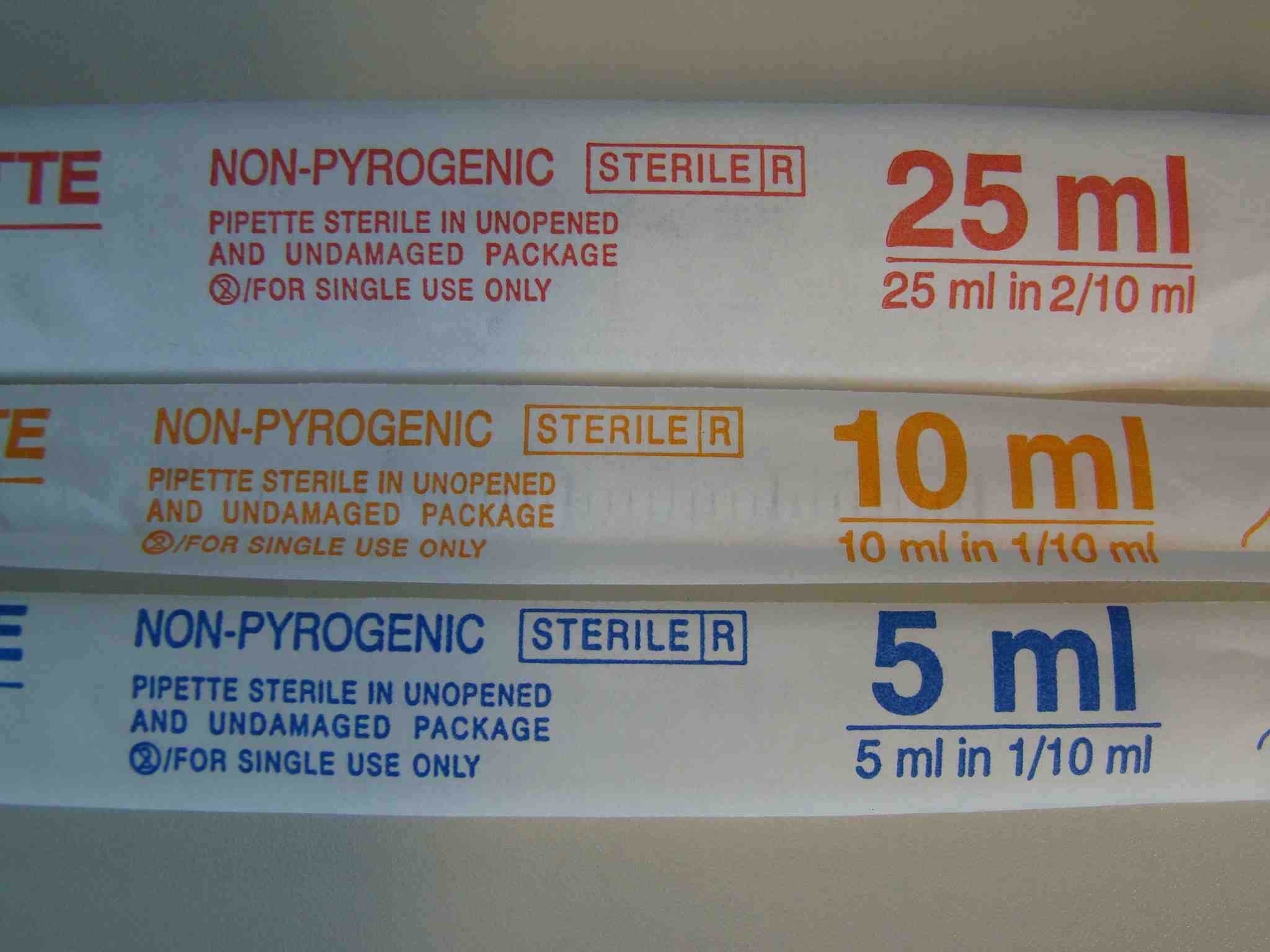
Please have a look to my demo of how to use them:
Settings of a typical serological pipette can be seen below. You can adjust the power of the pump that enables you to move volumes as small as 1/10 of ml. And you can switch between "drop wise" and "blow out" mode. You use "drop wise" mode when you do not want to disturb the cells on the bottom of a culturing dish.
[caption id="attachment_205" align="aligncenter" width="468" caption="head-of-serological-pipette"]
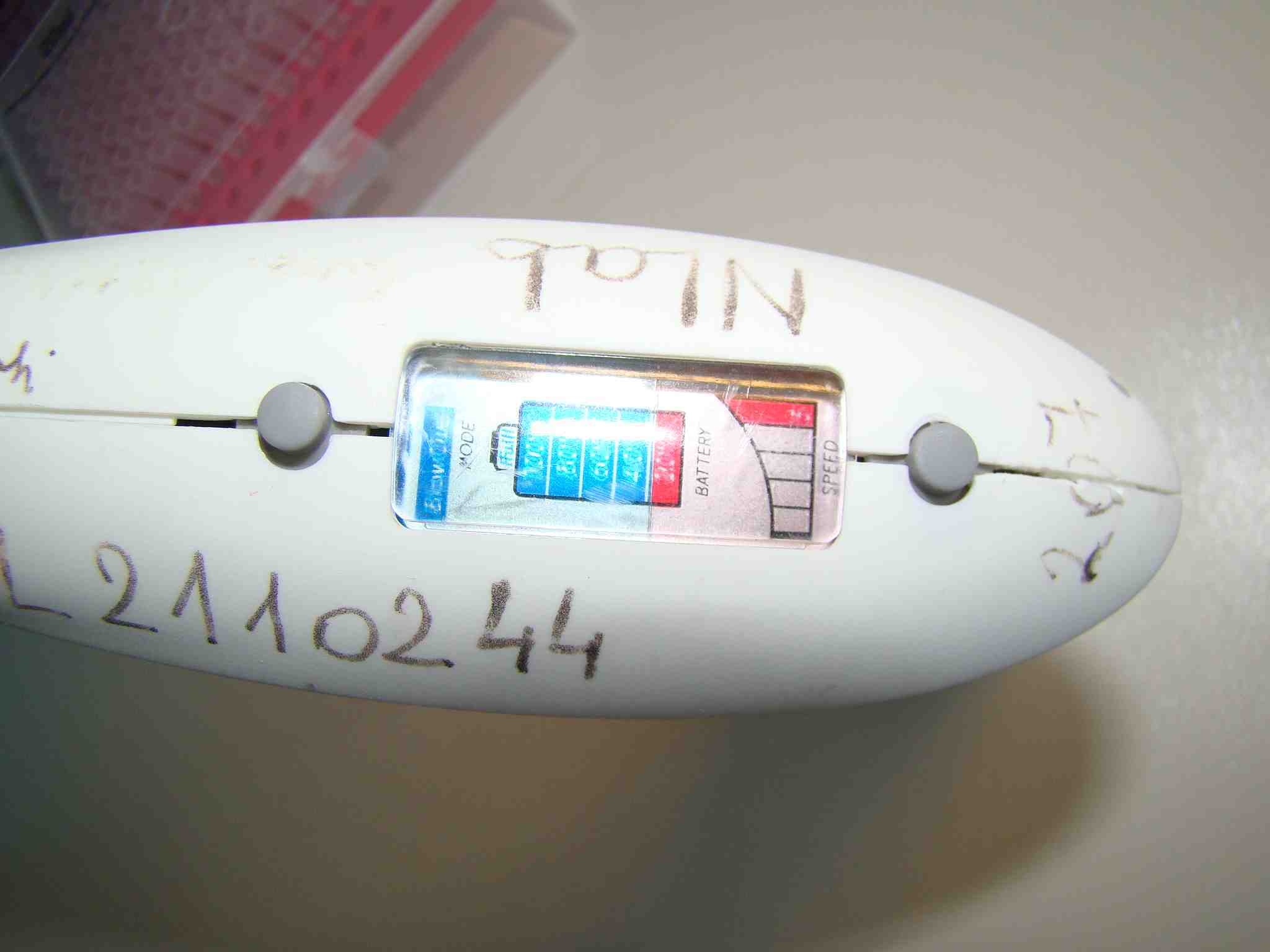 [/caption]
[/caption]A detailed demo about serological and other similar type of pipettes:
What about using Gilson type pipettes in the cell culture lab? We have sterile, pyrogenic free tips for the cell culture lab. This box contains for example certified DNAse, RNase and Pyrogen free tips. Each tip has an individual filter insert! You can use them for probably any protocoll in a standard molecular biology lab. Don't forget, they are not cheap at all...
[caption id="attachment_207" align="aligncenter" width="468" caption="barrier-tip-box-1ml"]
 [/caption]
[/caption][caption id="attachment_209" align="aligncenter" width="468" caption="barrier-tip-1ml"]
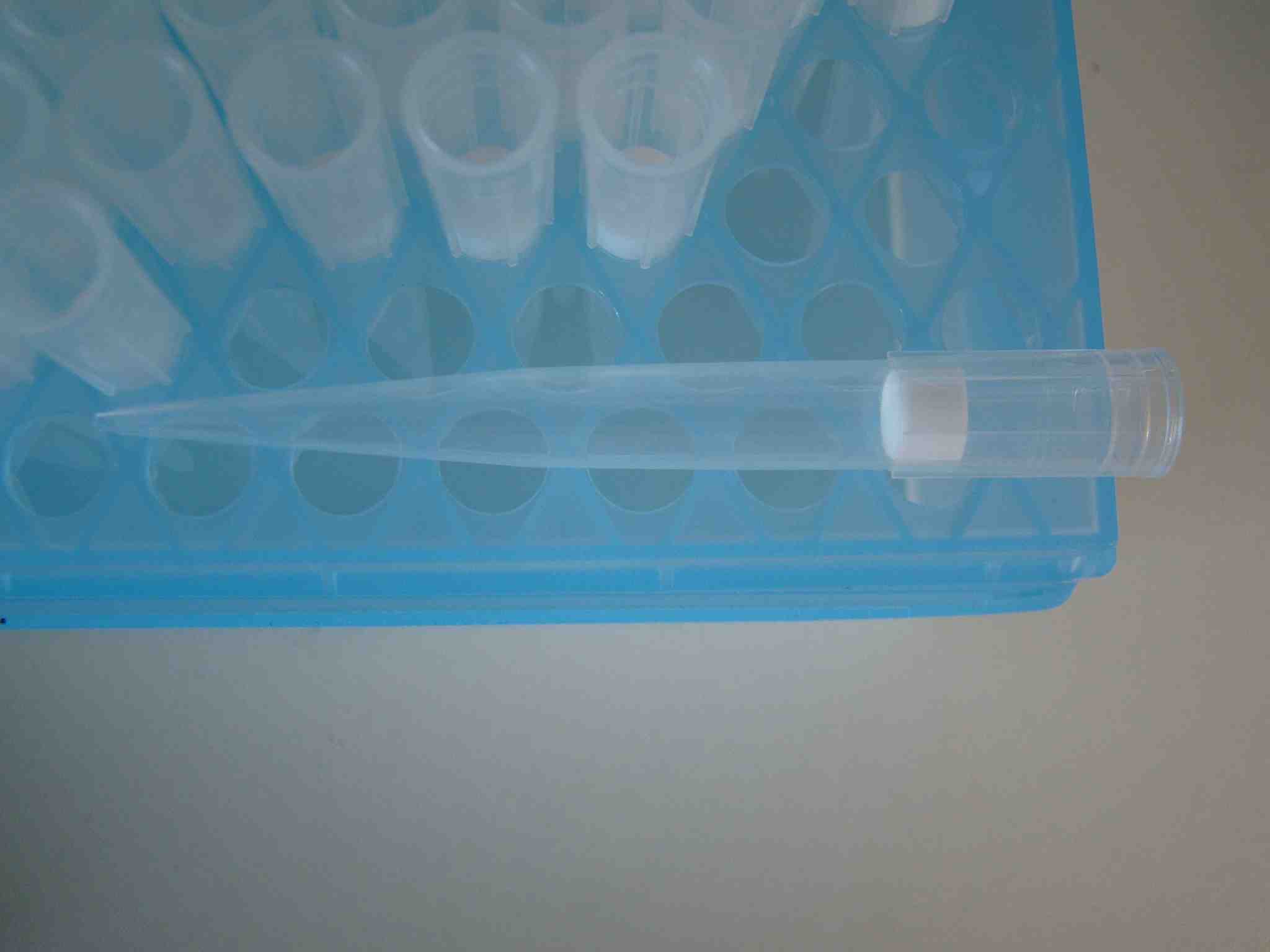 [/caption]
[/caption]Smaller volumes can be measured with smaller barrier tips:
[caption id="attachment_221" align="aligncenter" width="468" caption="barrier-tip-10ul"]
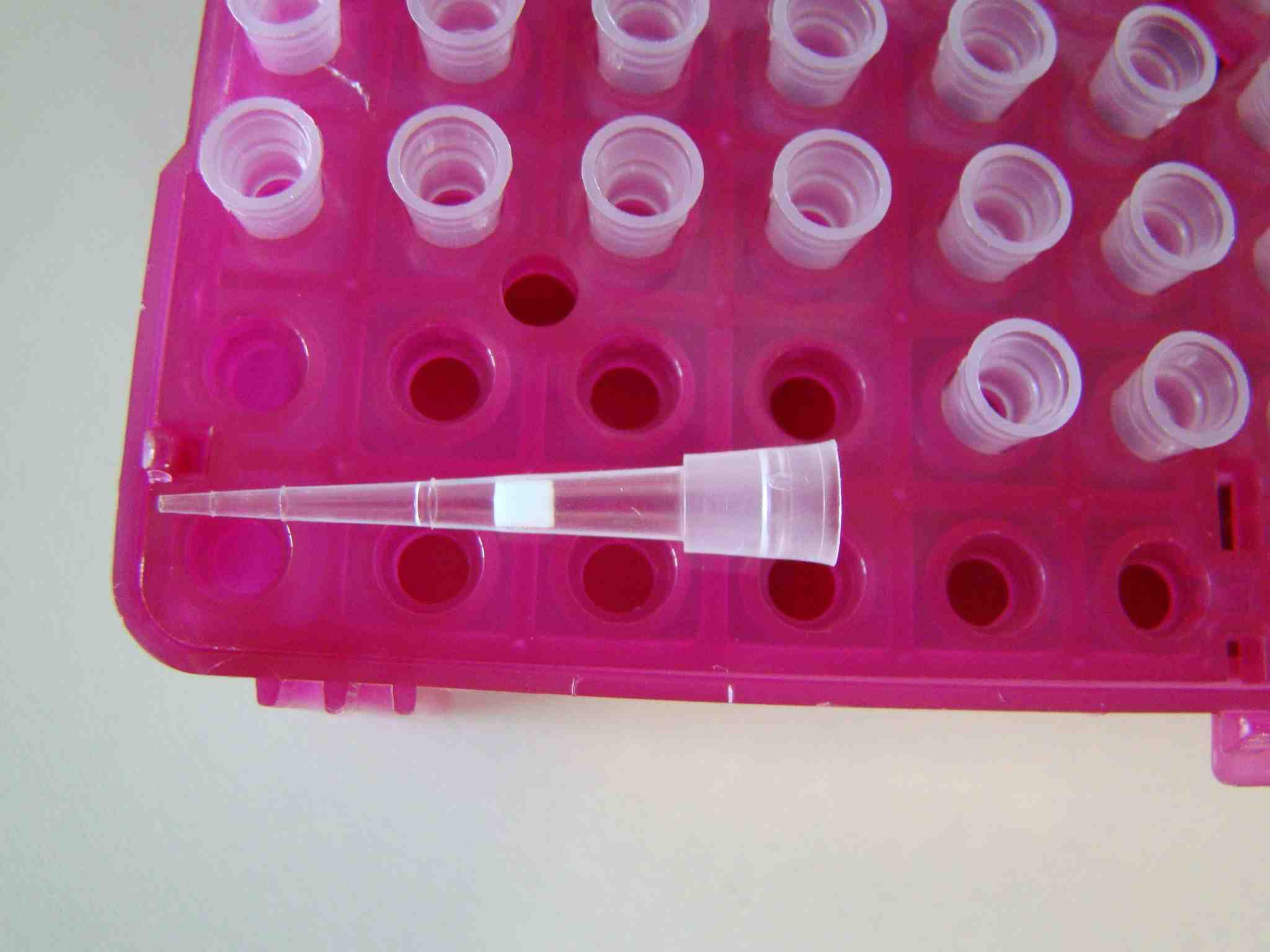 [/caption]
[/caption]I think we should go to the cell culture lab in one of our next post!
That's all for today, and let me have your feedback!
Plenty of good basic info regarding laboratory work is described <a href="">here.
Cheers,
Balint.
Subscribe to:
Posts (Atom)